The rticles package provides a suite of custom R Markdown LaTeX formats and templates for various formats. Most of the templates are provided and maintained by the community, and anyone can contribute a new template. See How to contribute below.
Installation
You can install and use rticles from CRAN as follows:
install.packages("rticles")If you wish to install the development version from GitHub (which often contains new article formats), you can do this:
# install.packages("pak")
pak::pak("rstudio/rticles")Using rticles
To use rticles from RStudio, you can access the templates through File -> New File -> R Markdown. This will open the dialog box where you can select from one of the available templates:
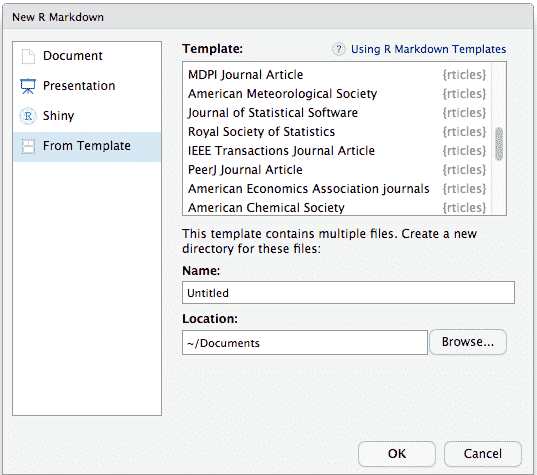
If you are not using RStudio, you’ll also need to install Pandoc following these instructions. Then, use the rmarkdown::draft() function to create articles:
rmarkdown::draft(
"MyJSSArticle.Rmd", template = "jss", package = "rticles"
)
rmarkdown::draft(
"MyRJournalArticle", template = "rjournal", package = "rticles"
)This will create a folder containing a Rmd file using the corresponding output format and all the assets required by this format.
Templates
Currently included templates and their contributors are the following:
You can also get the list of available journal names with rticles::journals().
rticles::journals()
#> [1] "acm" "acs" "aea" "agu"
#> [5] "ajs" "amq" "ams" "arxiv"
#> [9] "asa" "bioinformatics" "biometrics" "copernicus"
#> [13] "ctex" "elsevier" "frontiers" "glossa"
#> [17] "ieee" "ims" "informs" "iop"
#> [21] "isba" "jasa" "jedm" "joss"
#> [25] "jss" "lipics" "lncs" "mdpi"
#> [29] "mnras" "oup_v0" "oup_v1" "peerj"
#> [33] "pihph" "plos" "pnas" "rjournal"
#> [37] "rsos" "rss" "sage" "sim"
#> [41] "springer" "tf" "trb" "wellcomeor"Those are the values to use within rmarkdown::draft().
Under the hood, LaTeX templates are used to ensure that documents conform precisely to submission standards. At the same time, composition and formatting can be done using lightweight markdown syntax, and R code and its output can be seamlessly included using knitr.
Getting help
There are two main places to get help:
The RStudio community is a friendly place to ask any questions about rticles. Be sure to use the
rticlestag.Stack Overflow is a great source of answers to common bookdown questions. Use the tags
[r][rticles]if you ask a question.
Code of Conduct
Please note that the rticles project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
How to contribute?
Most of the templates are contributed directly by the users in the community. If you want rticles to offer a new journal format, you can contribute by the following way.
Suggest an idea for new format opening an issue.
You may not feel confident enough or may not have time to contribute a new format. By opening a new issue, you can share the idea for this format, and see if someone in the community can help on it.
This is not the best way to quickly get your format included but at least it is a great way to see if others are interested too.
To see the existing suggested formats, just filter issues with the help wanted ❤️ label. You can then add a 👍 or help to add the template 😉.
Contribute a new template format opening a pull request.
To contribute a new format, you need to open a new pull request (PR). When opening the PR, you’ll see the PR template explaining how to proceed and what is important to check. Please follow it.
Even if you are just starting or you are not finished, you share your work by creating a draft PR. It is a great way to let us know that you are still working on it (like these opened ones), and it is also a great way to ask for help from the community.
When you are ready, you can submit the PR for review, and we will iterate until it is merged.
Technical resources helpful to contribute a template
The best way to get started is to look at the previous examples of submitted PR. You’ll find links to them in the table above.
All the rticles format are build similarly by providing a new pandoc tex template to replace the default one. You’ll learn more about pandoc templates in these places:
You can study existing formats to see how all this works.

